Mahlon Collins
Researcher. Department of Genetics, Cell Biology, and Development @ University of Minnesota

I’m an interdisciplinary research with training in genomics, quantitative genetics, and neurobiology. I study protein homeostasis, the equilibrium of the abundance, activity, localization, folding, and turnover of cellular proteins. My work combines novel experimental approaches that I’ve developed with computational analyses of large-scale datasets. The long-term goal of my research is to understand the factors that regulate protein homeostasis and how changes in protein homeostasis contribute to variation in molecular, cellular, and organismal traits, including human health and disease.
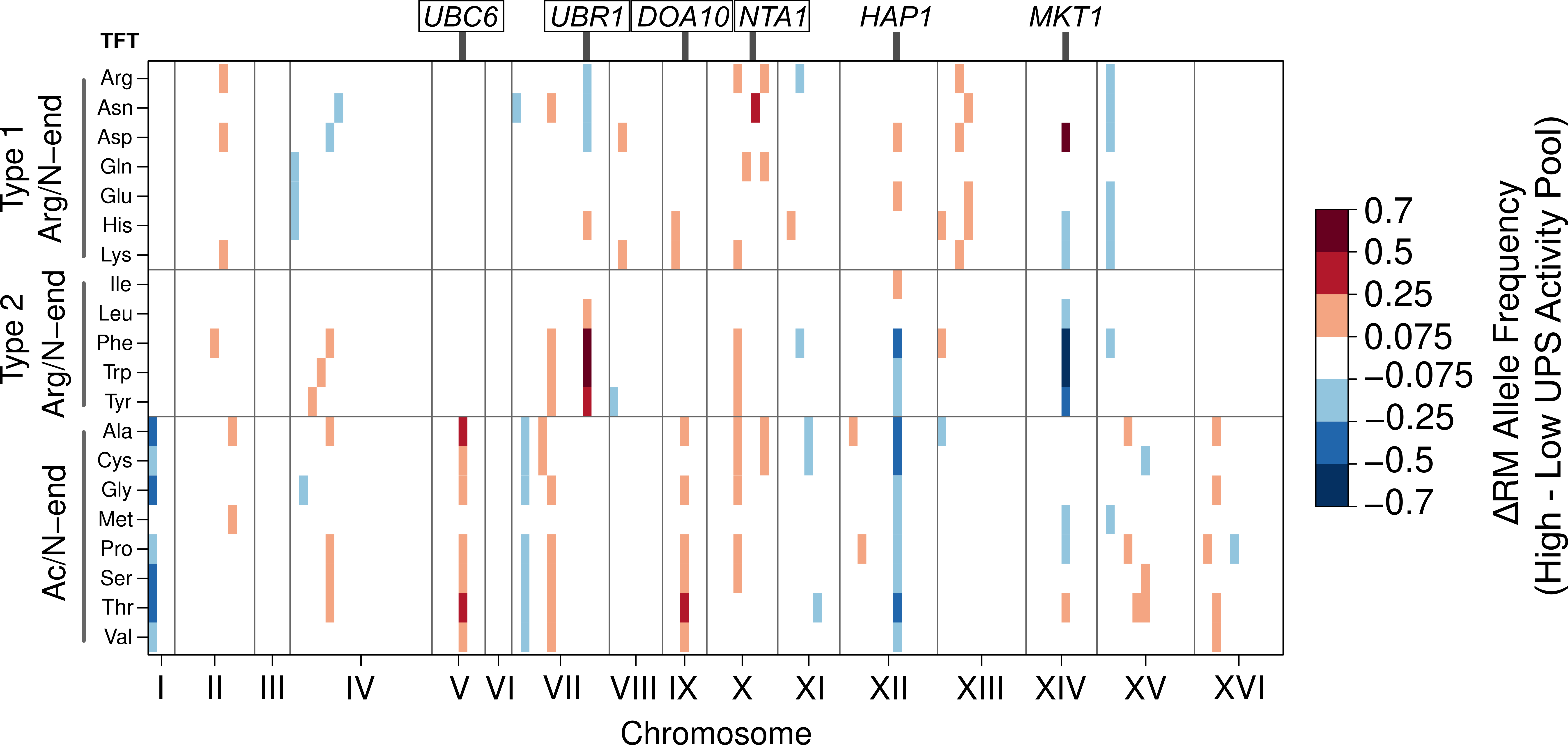 In my current work, I study how individual DNA sequence differences influence protein degradation. Protein degradation is an essential biological process that regulates protein abundance and removes misfolded and damaged proteins from cells. Through these actions, protein degradation regulates numerous aspects of cellular physiology, including gene expression, growth, development, and metabolism. However, prior to my recent work, we had almost no knowledge of how genetic variation affects the activity of protein degradation pathways or the degradation of individual proteins. I developed a novel, genome-wide approach for identifying genetic influences on protein degradation. My approach combines high-throughput synthetic reporters that allow me to measure protein degradation in millions of genetically variable single cells with a statistically powerful genetic mapping framework. This work provided the first genome-wide view of the genetics of protein degradation and identified multiple causal DNA variants that create pathway-specific changes in protein degradation.
In my current work, I study how individual DNA sequence differences influence protein degradation. Protein degradation is an essential biological process that regulates protein abundance and removes misfolded and damaged proteins from cells. Through these actions, protein degradation regulates numerous aspects of cellular physiology, including gene expression, growth, development, and metabolism. However, prior to my recent work, we had almost no knowledge of how genetic variation affects the activity of protein degradation pathways or the degradation of individual proteins. I developed a novel, genome-wide approach for identifying genetic influences on protein degradation. My approach combines high-throughput synthetic reporters that allow me to measure protein degradation in millions of genetically variable single cells with a statistically powerful genetic mapping framework. This work provided the first genome-wide view of the genetics of protein degradation and identified multiple causal DNA variants that create pathway-specific changes in protein degradation.
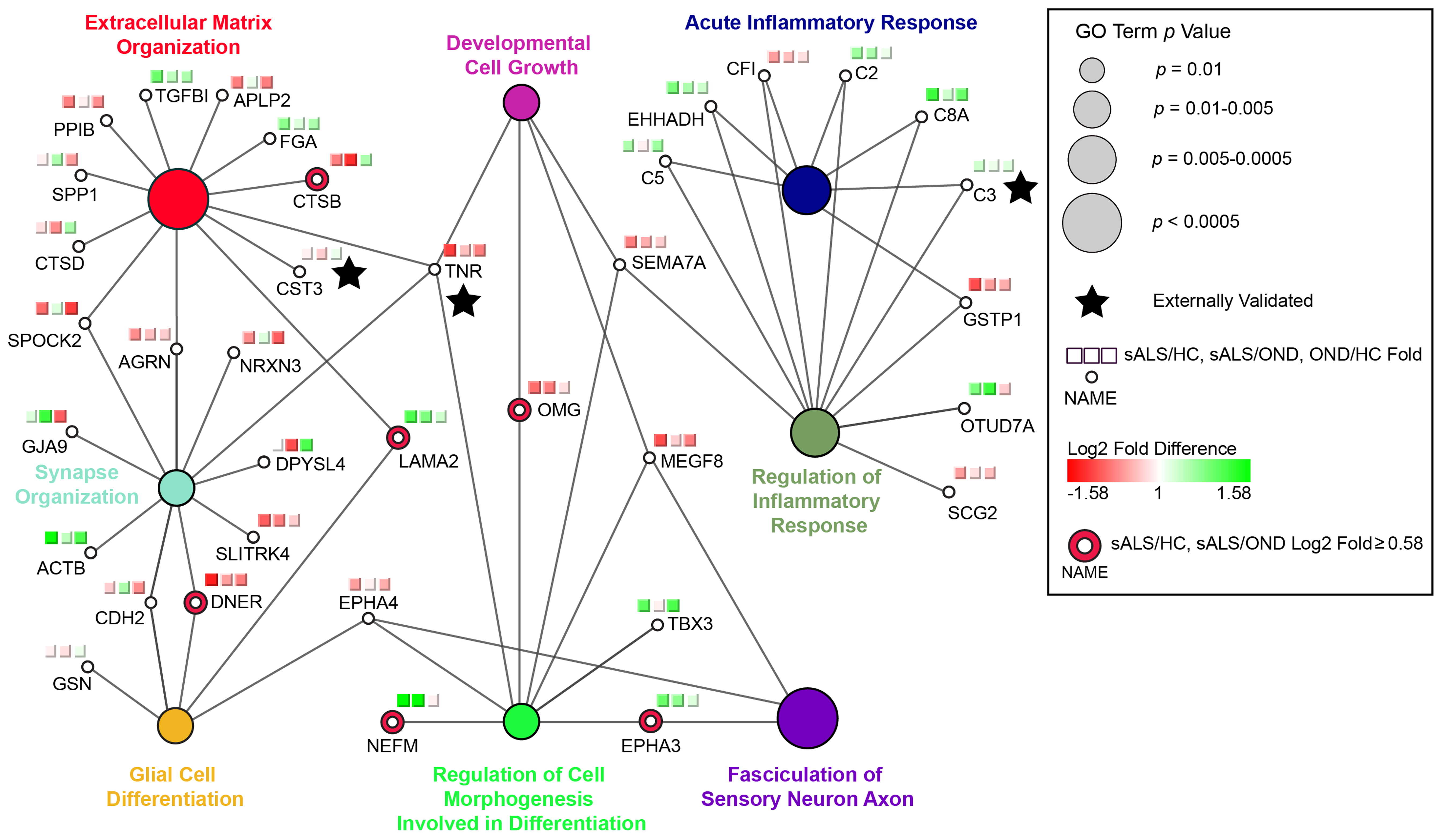 My prior work focused on identifying global proteomic changes in neurodegenerative diseases. I helped develop a new, scaled approach for mass-spectrometric proteomics of cerebrospinal fluid (CSF; the fluid surrounding the brain and spinal cord). I used the resulting datasets and machine learning to develop a sensitive, specific amyotrophic lateral sclerosis (ALS) protein biomarker panel. I also used CSF proteomic profiles to discover new neurodegenerative disease mechanisms. Using proteomic data, live-cell imaging, and image analysis, I discovered a novel mechanism by which RNA binding proteins form pathological protein aggregates in multiple neurodegenerative diseases, including ALS, Alzheimer’s disease, and dementia. Chronic entrapment of RNA binding proteins in stress-induced protein-RNA condensates (termed “nuclear stress bodies”), leads to the irreversible accumulation of RNA binding protein aggregates. This work expanded our understanding of the CSF proteome, identified novel ALS biomarkers, and identified nuclear stress bodies as a therapeutic target for multiple neurodegenerative disases.
My prior work focused on identifying global proteomic changes in neurodegenerative diseases. I helped develop a new, scaled approach for mass-spectrometric proteomics of cerebrospinal fluid (CSF; the fluid surrounding the brain and spinal cord). I used the resulting datasets and machine learning to develop a sensitive, specific amyotrophic lateral sclerosis (ALS) protein biomarker panel. I also used CSF proteomic profiles to discover new neurodegenerative disease mechanisms. Using proteomic data, live-cell imaging, and image analysis, I discovered a novel mechanism by which RNA binding proteins form pathological protein aggregates in multiple neurodegenerative diseases, including ALS, Alzheimer’s disease, and dementia. Chronic entrapment of RNA binding proteins in stress-induced protein-RNA condensates (termed “nuclear stress bodies”), leads to the irreversible accumulation of RNA binding protein aggregates. This work expanded our understanding of the CSF proteome, identified novel ALS biomarkers, and identified nuclear stress bodies as a therapeutic target for multiple neurodegenerative disases.
 I use GNU Emacs for many of my professional activities, such as writing, programming, data management, email, RSS, etc…., and write about how I’ve configured Emacs to meet my needs. My scientific work benefits especially from Emacs’s org-mode, ESS, and Ebib.
I use GNU Emacs for many of my professional activities, such as writing, programming, data management, email, RSS, etc…., and write about how I’ve configured Emacs to meet my needs. My scientific work benefits especially from Emacs’s org-mode, ESS, and Ebib.